PMA.start – universal whole slide image viewer for digital pathology
An end-user viewer and tile server in one convenient package
Download now for freeFor Windows, Linux and Mac
PMA.start runs on top of PMA.core.lite, which serves as a tile server.
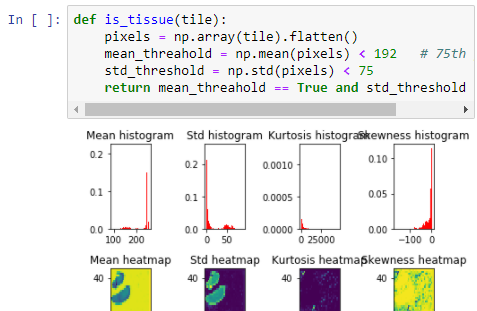
Utilizing our powerful webservice API and language-specific SDKs you can automate your image analysis workflows via scripting and use any image processing library you want.
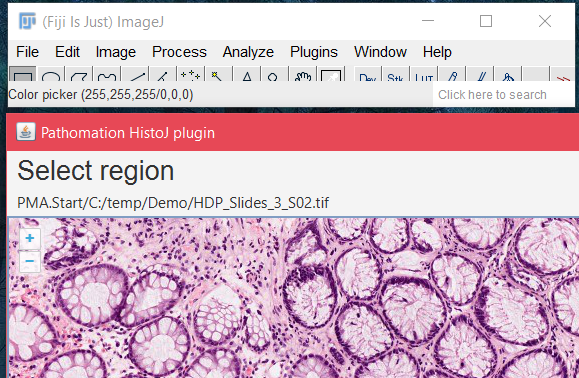
Through the ImageJ (FIJI) and QuPath plugins, you can bring any type of microscopic data into one of the most widely used image analysis programs on the market.
What do you get when you install PMA.start?
PMA.start is a free web based whole slide image viewer that:
A locally installed server application, built using Microsoft .Net framework, that:
An advanced tool to transfer slides from your local hard disk to your professional PMA.core tile server.
More information about PMA.transfer can be found on our main website.
A free command-line tool to convert any whole slide image to DICOM-compatible storage containers.
More information about our Dicomizer tool can be found at our RealData blog.
Our platform also makes use of various 3rd party software libraries all of which are listed here.